decoupler - Ensemble of methods to infer biological activities
decoupler is a package containing different statistical methods to extract biological activities from omics data within a unified framework. It allows to flexibly test any enrichment method with any prior knowledge resource and incorporates methods that take into account the sign and weight. It can be used with any omic, as long as its features can be linked to a biological process based on prior knowledge. For example, in transcriptomics gene sets regulated by a transcription factor, or in phospho-proteomics phosphosites that are targeted by a kinase.
This is its faster and memory efficient Python implementation, for the R version go here.
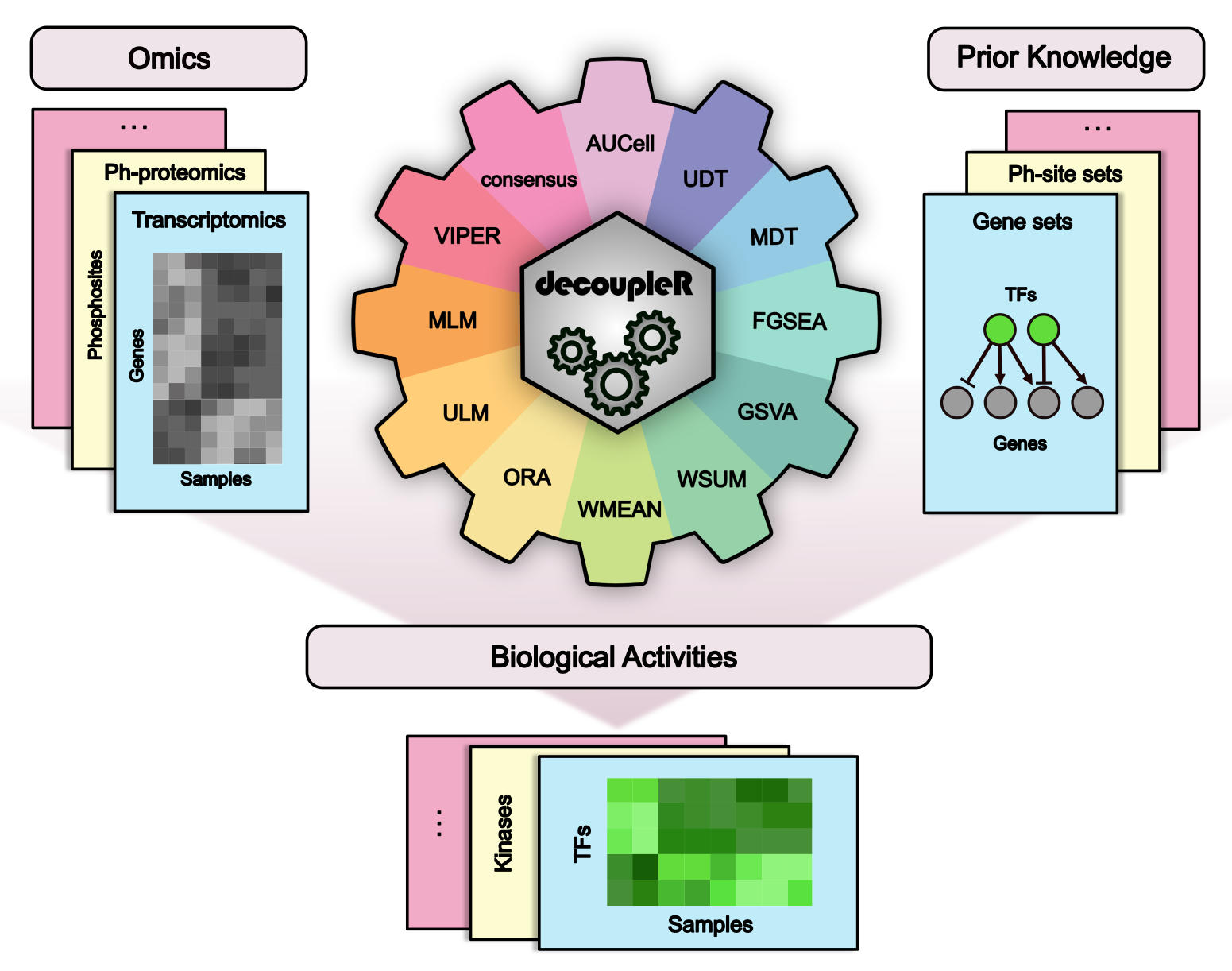
decoupler contains a collection of computational methods that coupled with prior knowledge resources estimate biological activities from omics data.
Check out the Usage or any other tutorial for further information.
If you have any question or problem do not hesitate to open an issue.
scverse
decoupler is part of the scverse ecosystem, a collection of tools for single-cell omics data analysis in python.
For more information check the link.
License
Footprint methods inside decoupler can be used for academic or commercial purposes, except viper which holds a non-commercial license.
The data redistributed by OmniPath does not have a license, each original resource carries their own. Here one can find the license information of all the resources in OmniPath.
Citation
Badia-i-Mompel P., Vélez Santiago J., Braunger J., Geiss C., Dimitrov D., Müller-Dott S., Taus P., Dugourd A., Holland C.H., Ramirez Flores R.O. and Saez-Rodriguez J. 2022. decoupleR: ensemble of computational methods to infer biological activities from omics data. Bioinformatics Advances. https://doi.org/10.1093/bioadv/vbac016

