Spatial functional analysis
Spatial transcriptomics technologies yield many molecular readouts that are hard to interpret by themselves. One way of summarizing this information is by inferring biological activities from prior knowledge.
In this notebook we showcase how to use decoupler for transcription factor and pathway activity inference from a human data-set. The data consists of a 10X Genomics Visium slide of a human lymph node and it is available at their website.
Note
This tutorial assumes that you already know the basics of decoupler. Else, check out the Usage tutorial first.
Loading packages
First, we need to load the relevant packages, scanpy to handle RNA-seq data and decoupler to use statistical methods.
[1]:
import scanpy as sc
import decoupler as dc
# Only needed for processing and plotting
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
Loading the data
We can download the data easily using scanpy:
[2]:
adata = sc.datasets.visium_sge(sample_id="V1_Human_Lymph_Node")
adata.var_names_make_unique()
adata
Variable names are not unique. To make them unique, call `.var_names_make_unique`.
Variable names are not unique. To make them unique, call `.var_names_make_unique`.
[2]:
AnnData object with n_obs × n_vars = 4035 × 36601
obs: 'in_tissue', 'array_row', 'array_col'
var: 'gene_ids', 'feature_types', 'genome'
uns: 'spatial'
obsm: 'spatial'
QC, projection and clustering
Here we follow the standard pre-processing steps as described in the scanpy vignette. These steps carry out the selection and filtration of spots based on quality control metrics, the data normalization and scaling, and the detection of highly variable features.
Note
This is just an example, these steps should change depending on the data.
[3]:
# Basic filtering
sc.pp.filter_cells(adata, min_genes=200)
sc.pp.filter_genes(adata, min_cells=10)
# Annotate the group of mitochondrial genes as 'mt'
adata.var['mt'] = adata.var_names.str.startswith('MT-')
sc.pp.calculate_qc_metrics(adata, qc_vars=['mt'], percent_top=None, log1p=False, inplace=True)
# Filter cells following standard QC criteria.
adata = adata[adata.obs.pct_counts_mt < 20, :]
# Normalize the data
sc.pp.normalize_total(adata, target_sum=1e4)
sc.pp.log1p(adata)
# Identify highly variable genes
sc.pp.highly_variable_genes(adata)
# Filter higly variable genes
adata.raw = adata
adata = adata[:, adata.var.highly_variable]
# Scale the data
sc.pp.scale(adata, max_value=10)
/home/badi/miniconda3/envs/decoupler/lib/python3.8/site-packages/scanpy/preprocessing/_normalization.py:155: UserWarning: Revieved a view of an AnnData. Making a copy.
view_to_actual(adata)
/home/badi/miniconda3/envs/decoupler/lib/python3.8/site-packages/scanpy/preprocessing/_simple.py:843: UserWarning: Revieved a view of an AnnData. Making a copy.
view_to_actual(adata)
Then we group spots based on the similarity of their transcription profiles. We can visualize the obtianed clusters on a UMAP or directly at the tissue:
[4]:
# Generate PCA features
sc.tl.pca(adata, svd_solver='arpack')
# Compute distances in the PCA space, and find spot neighbors
sc.pp.neighbors(adata)
# Run leiden clustering algorithm
sc.tl.leiden(adata)
# Visualize
sc.pl.spatial(adata, color=[None, 'leiden'], size=1.5, wspace=0)
/home/badi/miniconda3/envs/decoupler/lib/python3.8/site-packages/anndata/_core/anndata.py:1220: FutureWarning: The `inplace` parameter in pandas.Categorical.reorder_categories is deprecated and will be removed in a future version. Reordering categories will always return a new Categorical object.
c.reorder_categories(natsorted(c.categories), inplace=True)
... storing 'feature_types' as categorical
/home/badi/miniconda3/envs/decoupler/lib/python3.8/site-packages/anndata/_core/anndata.py:1220: FutureWarning: The `inplace` parameter in pandas.Categorical.reorder_categories is deprecated and will be removed in a future version. Reordering categories will always return a new Categorical object.
c.reorder_categories(natsorted(c.categories), inplace=True)
... storing 'genome' as categorical
Transcription factor activity inference
The first functional analysis we can perform is to infer transcription factor (TF) activities from our transcriptomics data. We will need a gene regulatory network (GRN) and a statistical method.
DoRothEA network
DoRothEA is a comprehensive resource containing a curated collection of TFs and their transcriptional targets. Since these regulons were gathered from different types of evidence, interactions in DoRothEA are classified in different confidence levels, ranging from A (highest confidence) to D (lowest confidence). Moreover, each interaction is weighted by its confidence level and the sign of its mode of regulation (activation or inhibition).
For this example we will use the human version (other organisms are available) and we will use the confidence levels ABC. To access it we can use decoupler.
Note
In this tutorial we use the network DoRothEA, but we could use any other GRN coming from an inference method such as CellOracle, pySCENIC or SCENIC+.
[5]:
net = dc.get_dorothea(organism='human', levels=['A','B','C'])
net
[5]:
| source | confidence | target | weight | |
|---|---|---|---|---|
| 0 | ETS1 | A | IL12B | 1.000000 |
| 1 | RELA | A | IL6 | 1.000000 |
| 2 | MITF | A | BCL2A1 | -1.000000 |
| 3 | E2F1 | A | TRERF1 | 1.000000 |
| 4 | MITF | A | BCL2 | 1.000000 |
| ... | ... | ... | ... | ... |
| 32272 | IKZF1 | C | PTK2B | 0.333333 |
| 32273 | IKZF1 | C | PRKCB | 0.333333 |
| 32274 | IKZF1 | C | PREX1 | 0.333333 |
| 32275 | IRF4 | C | SLAMF7 | 0.333333 |
| 32276 | ZNF83 | C | ZNF331 | 0.333333 |
32277 rows × 4 columns
Activity inference with Multivariate Linear Model
To infer activities we will run the Multivariate Linear Model method (mlm). It models the observed gene expression by using a regulatory adjacency matrix (target genes x TFs) as covariates of a linear model. The values of this matrix are the associated interaction weights. The obtained t-values of the fitted model are the activity scores.
To run decoupler methods, we need an input matrix (mat), an input prior knowledge network/resource (net), and the name of the columns of net that we want to use.
[6]:
dc.run_mlm(mat=adata, net=net, source='source', target='target', weight='weight', verbose=True)
Running mlm on mat with 4032 samples and 19813 targets for 294 sources.
100%|██████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████| 1/1 [00:06<00:00, 6.84s/it]
The obtained scores (t-values)(mlm_estimate) and p-values (mlm_pvals) are stored in the .obsm key:
[7]:
adata.obsm['mlm_estimate']
[7]:
| AHR | AR | ARID2 | ARID3A | ARNT | ARNTL | ASCL1 | ATF1 | ATF2 | ATF3 | ... | ZNF217 | ZNF24 | ZNF263 | ZNF274 | ZNF384 | ZNF584 | ZNF592 | ZNF639 | ZNF644 | ZNF740 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AAACAAGTATCTCCCA-1 | 0.531388 | 4.314746 | 2.580497 | -2.264766 | -0.211907 | 0.031246 | -0.705776 | -0.118562 | 0.799197 | -0.045215 | ... | 0.618186 | -0.924915 | -1.565854 | -0.710968 | 1.119455 | 0.527442 | 0.561804 | 3.302261 | -0.169674 | 1.063122 |
| AAACAATCTACTAGCA-1 | -0.684831 | 3.082588 | 2.525492 | -0.760523 | -0.134987 | -1.309098 | -0.090339 | -0.262004 | 1.189955 | -0.133106 | ... | 0.001439 | -1.933857 | -1.526967 | -0.431441 | 0.914302 | 0.337622 | 1.228477 | 3.548201 | -0.341155 | 0.854290 |
| AAACACCAATAACTGC-1 | -0.966583 | 3.456383 | 0.965503 | -2.124058 | -0.092816 | -1.105180 | -0.122935 | 1.216181 | 1.331234 | 0.694221 | ... | 1.274752 | -1.129040 | -1.979256 | 0.094362 | 1.624496 | 0.581281 | -0.018216 | 2.823706 | -0.299057 | 0.781646 |
| AAACAGAGCGACTCCT-1 | 0.084335 | 3.552887 | 2.330085 | -1.771000 | -0.805615 | -0.930001 | -0.450489 | 0.392044 | 1.208560 | -0.174565 | ... | 0.172208 | -1.423535 | -1.436785 | -1.608596 | 1.548968 | 0.492690 | 0.748371 | 2.668740 | 1.290589 | 0.898669 |
| AAACAGCTTTCAGAAG-1 | -0.113021 | 3.062887 | 1.692873 | -1.376210 | -0.237199 | -1.540030 | 0.218669 | 0.059750 | 1.091377 | -0.090121 | ... | 0.990816 | -1.068140 | -1.429415 | -0.121686 | 0.748879 | 0.354986 | 0.187215 | 2.666389 | 0.885567 | 0.440206 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| TTGTTTCACATCCAGG-1 | -0.890853 | 3.760496 | 1.341087 | -2.143296 | 0.199219 | -1.063316 | 0.574174 | -0.232284 | -0.531052 | -0.697277 | ... | 2.323004 | -1.538032 | -2.160935 | -0.829605 | 0.601897 | -0.741982 | 0.184341 | 2.963787 | -0.056685 | 0.354788 |
| TTGTTTCATTAGTCTA-1 | 1.157550 | 3.614088 | 0.934339 | -1.603928 | -1.034015 | -1.440529 | -0.260510 | -0.934655 | 0.368782 | 0.272667 | ... | 1.935458 | -1.350134 | -1.803886 | -1.309194 | 0.239783 | 0.030232 | -0.161986 | 3.265780 | 0.185732 | 2.117301 |
| TTGTTTCCATACAACT-1 | -1.258395 | 2.891093 | 3.206188 | -1.074143 | -0.382113 | -0.945535 | -0.151498 | 1.225757 | 1.193222 | -0.135125 | ... | 1.379948 | -0.543945 | -1.674882 | -0.879069 | 1.404226 | 0.835917 | -0.477097 | 2.610413 | -0.327709 | 1.427865 |
| TTGTTTGTATTACACG-1 | -0.739230 | 2.823546 | 3.534382 | -1.066094 | 0.324376 | -1.311501 | -1.026220 | 0.729384 | 1.583481 | 0.750026 | ... | 0.692039 | -1.517446 | -1.793239 | -0.583169 | 1.176431 | 0.302630 | -0.199994 | 2.923765 | -1.083028 | 0.244638 |
| TTGTTTGTGTAAATTC-1 | 0.159324 | 3.061946 | 2.349993 | -0.897393 | 0.464201 | -1.336176 | -0.436003 | -0.506391 | -0.004222 | 0.715258 | ... | 0.998253 | -1.181231 | -0.979263 | -1.144093 | 1.208452 | 0.733775 | 0.272451 | 3.959256 | 1.771204 | 1.412840 |
4032 rows × 294 columns
Note: Each run of run_mlm overwrites what is inside of mlm_estimate and mlm_pvals. if you want to run mlm with other resources and still keep the activities inside the same AnnData object, you can store the results in any other key in .obsm with different names, for example:
[8]:
adata.obsm['dorothea_mlm_estimate'] = adata.obsm['mlm_estimate'].copy()
adata.obsm['dorothea_mlm_pvals'] = adata.obsm['mlm_pvals'].copy()
adata
[8]:
AnnData object with n_obs × n_vars = 4032 × 2491
obs: 'in_tissue', 'array_row', 'array_col', 'n_genes', 'n_genes_by_counts', 'total_counts', 'total_counts_mt', 'pct_counts_mt', 'leiden'
var: 'gene_ids', 'feature_types', 'genome', 'n_cells', 'mt', 'n_cells_by_counts', 'mean_counts', 'pct_dropout_by_counts', 'total_counts', 'highly_variable', 'means', 'dispersions', 'dispersions_norm', 'mean', 'std'
uns: 'spatial', 'log1p', 'hvg', 'pca', 'neighbors', 'leiden', 'leiden_colors'
obsm: 'spatial', 'X_pca', 'mlm_estimate', 'mlm_pvals', 'dorothea_mlm_estimate', 'dorothea_mlm_pvals'
varm: 'PCs'
obsp: 'distances', 'connectivities'
Visualization
To visualize the obtained scores, we can re-use many of scanpy’s plotting functions. First though, we need to extract them from the adata object.
[9]:
acts = dc.get_acts(adata, obsm_key='dorothea_mlm_estimate')
acts
[9]:
AnnData object with n_obs × n_vars = 4032 × 294
obs: 'in_tissue', 'array_row', 'array_col', 'n_genes', 'n_genes_by_counts', 'total_counts', 'total_counts_mt', 'pct_counts_mt', 'leiden'
uns: 'spatial', 'log1p', 'hvg', 'pca', 'neighbors', 'leiden', 'leiden_colors'
obsm: 'spatial', 'X_pca', 'mlm_estimate', 'mlm_pvals', 'dorothea_mlm_estimate', 'dorothea_mlm_pvals'
dc.get_acts returns a new AnnData object which holds the obtained activities in its .X attribute, allowing us to re-use many scanpy functions, for example:
[10]:
sc.pl.spatial(acts, color=['leiden', 'PAX5'], cmap='coolwarm', size=1.5, wspace=0)
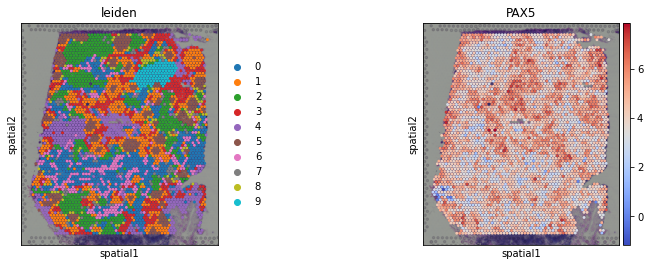
Here we observe the activity infered for PAX5 across spots. Interestingly, PAX5 is a known TF crucial for B cell identity and function. The inference of activities from “foot-prints” of target genes is more informative than just looking at the molecular readouts of a given TF, as an example here is the gene expression of PAX5, which is not very informative by itself:
[11]:
sc.pl.spatial(adata, color='PAX5', size=1.5)

Exploration
With decoupler we can also see what is the mean activity per group (to show more TFs, increase the min_std parameter):
[12]:
mean_acts = dc.summarize_acts(acts, groupby='leiden', min_std=0.4)
mean_acts
[12]:
| ATF6 | E2F4 | ERG | ETS1 | IRF1 | IRF9 | JUNB | MYC | PAX5 | RBPJ | RFX5 | STAT1 | STAT2 | STAT3 | TBX21 | TFDP1 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 5.425647 | 5.950534 | 0.976629 | 7.022013 | -0.296379 | 0.364462 | 1.528090 | 13.706879 | 3.980209 | -0.702463 | 11.314269 | 1.457677 | 3.198416 | 2.211520 | 1.840758 | 1.651450 |
| 1 | 3.962387 | 6.329461 | 0.899010 | 7.067654 | -0.045027 | 0.298553 | 2.174330 | 14.132327 | 5.419888 | 0.276466 | 12.545614 | 1.347172 | 3.390375 | 1.564702 | 1.688480 | 1.655355 |
| 2 | 4.308863 | 6.204994 | 0.908421 | 7.663957 | -0.114513 | 0.257776 | 1.688271 | 14.019668 | 3.838306 | -0.979750 | 10.227130 | 1.548294 | 3.774739 | 1.867868 | 2.608472 | 1.564099 |
| 3 | 4.677002 | 6.115650 | 1.481284 | 7.245724 | -0.132964 | 0.336158 | 1.832213 | 14.238405 | 4.562850 | -0.398871 | 12.552104 | 1.873078 | 3.420067 | 2.550702 | 1.399548 | 1.762380 |
| 4 | 4.512222 | 5.423492 | 1.353292 | 6.270385 | -0.105885 | 0.345371 | 1.903540 | 13.461339 | 4.112409 | -0.179700 | 11.339410 | 1.291094 | 2.708822 | 2.374198 | 1.275249 | 1.746006 |
| 5 | 3.962252 | 9.133594 | 0.696790 | 6.748394 | -0.343900 | 0.270207 | 2.835591 | 14.928737 | 5.408315 | 0.556190 | 12.102087 | 0.981099 | 2.347938 | 1.259365 | 1.193647 | 4.038791 |
| 6 | 5.333444 | 5.908932 | 1.525536 | 7.519007 | -0.266074 | 0.298177 | 1.360018 | 14.022554 | 4.233101 | -0.598897 | 12.145805 | 1.924856 | 3.262287 | 2.700020 | 1.615572 | 1.588291 |
| 7 | 4.924606 | 6.188310 | 1.961460 | 7.522125 | -0.171967 | 0.355779 | 1.561548 | 14.214684 | 3.937887 | -0.789719 | 11.407527 | 1.688864 | 3.329131 | 2.477357 | 1.921157 | 1.785569 |
| 8 | 4.742176 | 7.305005 | 0.868916 | 7.338912 | -0.117163 | 0.395704 | 1.915956 | 14.851122 | 4.743230 | -0.567329 | 11.423132 | 1.358365 | 3.459237 | 1.808232 | 1.838801 | 2.466387 |
| 9 | 4.523947 | 6.937476 | 0.825905 | 7.283448 | 1.095696 | 1.769770 | 1.874522 | 14.956728 | 4.442098 | -0.750603 | 10.470515 | 2.704625 | 10.020187 | 1.340084 | 2.119819 | 2.382874 |
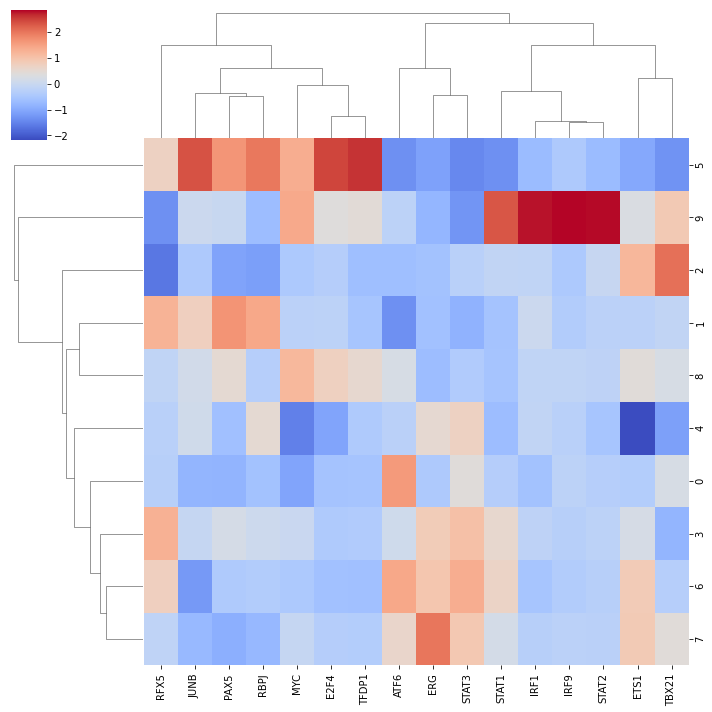
We can visualize which TF is more active across clusters using seaborn:
[13]:
sns.clustermap(mean_acts, xticklabels=mean_acts.columns, cmap='coolwarm', z_score=1)
plt.show()
We can visualize again on the tissue some marker TFs:
[14]:
sc.pl.spatial(acts, color=['leiden', 'IRF1', 'TFDP1', 'STAT3'], cmap='coolwarm', size=1.5, wspace=0)

Pathway activity inference
We can also infer pathway activities from our transcriptomics data.
PROGENy model
PROGENy is a comprehensive resource containing a curated collection of pathways and their target genes, with weights for each interaction. For this example we will use the human weights (other organisms are available) and we will use the top 100 responsive genes ranked by p-value. Here is a brief description of each pathway:
Androgen: involved in the growth and development of the male reproductive organs.
EGFR: regulates growth, survival, migration, apoptosis, proliferation, and differentiation in mammalian cells
Estrogen: promotes the growth and development of the female reproductive organs.
Hypoxia: promotes angiogenesis and metabolic reprogramming when O2 levels are low.
JAK-STAT: involved in immunity, cell division, cell death, and tumor formation.
MAPK: integrates external signals and promotes cell growth and proliferation.
NFkB: regulates immune response, cytokine production and cell survival.
p53: regulates cell cycle, apoptosis, DNA repair and tumor suppression.
PI3K: promotes growth and proliferation.
TGFb: involved in development, homeostasis, and repair of most tissues.
TNFa: mediates haematopoiesis, immune surveillance, tumour regression and protection from infection.
Trail: induces apoptosis.
VEGF: mediates angiogenesis, vascular permeability, and cell migration.
WNT: regulates organ morphogenesis during development and tissue repair.
To access it we can use decoupler.
[15]:
model = dc.get_progeny(organism='human', top=100)
model
[15]:
| source | target | weight | p_value | |
|---|---|---|---|---|
| 0 | Androgen | TMPRSS2 | 11.490631 | 0.000000e+00 |
| 1 | Androgen | NKX3-1 | 10.622551 | 2.242078e-44 |
| 2 | Androgen | MBOAT2 | 10.472733 | 4.624285e-44 |
| 3 | Androgen | KLK2 | 10.176186 | 1.944414e-40 |
| 4 | Androgen | SARG | 11.386852 | 2.790209e-40 |
| ... | ... | ... | ... | ... |
| 1395 | p53 | CCDC150 | -3.174527 | 7.396252e-13 |
| 1396 | p53 | LCE1A | 6.154823 | 8.475458e-13 |
| 1397 | p53 | TREM2 | 4.101937 | 9.739648e-13 |
| 1398 | p53 | GDF9 | 3.355741 | 1.087433e-12 |
| 1399 | p53 | NHLH2 | 2.201638 | 1.651582e-12 |
1400 rows × 4 columns
Activity inference with Multivariate Linear Model
Like in the previous section, we will use mlm to infer activities:
[16]:
dc.run_mlm(mat=adata, net=model, source='source', target='target', weight='weight', verbose=True)
# Store them in a different key
adata.obsm['progeny_mlm_estimate'] = adata.obsm['mlm_estimate'].copy()
adata.obsm['progeny_mlm_pvals'] = adata.obsm['mlm_pvals'].copy()
Running mlm on mat with 4032 samples and 19813 targets for 14 sources.
100%|██████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████| 1/1 [00:04<00:00, 4.79s/it]
The obtained scores (t-values)(mlm_estimate) and p-values (mlm_pvals) are stored in the .obsm key:
[17]:
adata.obsm['progeny_mlm_estimate']
[17]:
| Androgen | EGFR | Estrogen | Hypoxia | JAK-STAT | MAPK | NFkB | PI3K | TGFb | TNFa | Trail | VEGF | WNT | p53 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AAACAAGTATCTCCCA-1 | -0.188486 | 1.208809 | 0.845334 | 2.824020 | 2.956565 | 0.844544 | -1.375955 | -1.658008 | -0.506245 | 3.400028 | 0.948775 | 0.354302 | -0.394136 | -0.592207 |
| AAACAATCTACTAGCA-1 | -0.296953 | -0.593280 | -0.558594 | 1.577488 | 3.994631 | -0.434184 | -0.145056 | -2.026974 | -1.356935 | 2.522995 | 0.198127 | 0.449313 | -0.742038 | 0.015973 |
| AAACACCAATAACTGC-1 | -0.438746 | 0.179741 | 0.412451 | 1.812180 | 4.110078 | 0.803520 | -1.396133 | -2.972383 | -1.486423 | 4.421825 | 0.567402 | 0.804890 | -0.735639 | 0.174948 |
| AAACAGAGCGACTCCT-1 | 0.007000 | 0.652227 | -0.821658 | 2.101911 | 22.972557 | -0.683112 | -1.704974 | -1.953475 | -1.653204 | 3.975250 | 1.694650 | 0.755143 | -0.501892 | 0.129601 |
| AAACAGCTTTCAGAAG-1 | -0.482217 | 0.393672 | -0.576826 | 1.959044 | 4.177846 | -0.527857 | -0.996393 | -3.752962 | -1.633997 | 2.832229 | 0.967923 | -0.093649 | -0.575737 | 0.212081 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| TTGTTTCACATCCAGG-1 | -0.292770 | 1.405961 | 0.330157 | 1.788581 | 4.066021 | -0.854148 | -1.628868 | -2.403106 | -1.488374 | 3.821350 | 0.888249 | 0.231786 | -0.673330 | -0.536630 |
| TTGTTTCATTAGTCTA-1 | 0.216849 | 0.954299 | -0.934332 | 2.779557 | 3.680631 | 0.638986 | -0.052738 | -1.530679 | -1.766615 | 3.159299 | -0.192867 | 0.422701 | -0.358662 | 0.280121 |
| TTGTTTCCATACAACT-1 | -0.351691 | 0.822820 | 0.292143 | 1.386598 | 3.325152 | 0.149769 | -0.320018 | -2.387814 | -0.621196 | 3.825063 | 0.242561 | -0.025989 | 0.193019 | 0.199643 |
| TTGTTTGTATTACACG-1 | -1.506403 | 0.693628 | -0.210152 | 2.470843 | 4.737563 | -0.181501 | -0.572574 | -1.724866 | -0.646039 | 2.713047 | -0.016597 | 0.299411 | -0.704308 | -0.432493 |
| TTGTTTGTGTAAATTC-1 | -0.116604 | 0.914778 | -0.572922 | 1.167968 | 4.379010 | -1.397615 | -0.151022 | -2.901103 | -2.101318 | 1.985720 | 0.722871 | 0.648947 | -1.590521 | 0.610262 |
4032 rows × 14 columns
Visualization
Like in the previous section, we will extract the activities from the adata object.
[18]:
acts = dc.get_acts(adata, obsm_key='progeny_mlm_estimate')
acts
[18]:
AnnData object with n_obs × n_vars = 4032 × 14
obs: 'in_tissue', 'array_row', 'array_col', 'n_genes', 'n_genes_by_counts', 'total_counts', 'total_counts_mt', 'pct_counts_mt', 'leiden'
uns: 'spatial', 'log1p', 'hvg', 'pca', 'neighbors', 'leiden', 'leiden_colors'
obsm: 'spatial', 'X_pca', 'mlm_estimate', 'mlm_pvals', 'dorothea_mlm_estimate', 'dorothea_mlm_pvals', 'progeny_mlm_estimate', 'progeny_mlm_pvals'
Once extracted we can visualize them:
[19]:
sc.pl.spatial(acts, color='Trail', cmap='coolwarm', size=1.5)

Here we show the activity of the pathway Trail, which is associated with apoptosis.
Exploration
Let us see the mean pathway activity per cluster:
[20]:
mean_acts = dc.summarize_acts(acts, groupby='leiden', min_std=0)
mean_acts
[20]:
| Androgen | EGFR | Estrogen | Hypoxia | JAK-STAT | MAPK | NFkB | PI3K | TGFb | TNFa | Trail | VEGF | WNT | p53 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | -0.235733 | 0.390685 | 0.057316 | 2.028656 | 3.310778 | -0.082057 | -0.693550 | -2.151323 | -1.103416 | 2.876699 | 0.877714 | 0.212958 | -0.744617 | 0.018514 |
| 1 | -0.361732 | 0.395693 | -0.508569 | 1.789956 | 3.559256 | -0.339410 | -0.866924 | -2.090730 | -1.236661 | 2.921738 | 1.268237 | 0.484229 | -0.683389 | -0.038626 |
| 2 | -0.376320 | 0.267465 | -0.413390 | 2.016582 | 3.970872 | -0.114459 | -0.707642 | -2.385279 | -1.294334 | 3.097718 | 0.508334 | 0.561026 | -0.735835 | 0.041692 |
| 3 | -0.247242 | 0.840521 | -0.134884 | 2.248527 | 3.830161 | -0.104251 | -0.472488 | -2.093627 | -0.693436 | 3.823169 | 1.041569 | 0.493589 | -0.611709 | 0.063977 |
| 4 | -0.262289 | 0.643110 | 0.029117 | 1.830205 | 3.153491 | -0.335399 | -0.823879 | -1.951632 | -0.444595 | 3.039439 | 1.043293 | 0.136829 | -0.619363 | -0.133458 |
| 5 | 0.055557 | 0.533585 | -0.446169 | 1.856608 | 2.214398 | -0.528827 | -1.073485 | -1.667952 | -1.513876 | 2.833984 | 1.222594 | 0.461531 | -0.993194 | -0.156339 |
| 6 | -0.157354 | 0.582798 | 0.130340 | 2.260429 | 3.516362 | 0.081085 | -0.560231 | -2.286241 | -0.858187 | 3.446463 | 0.952809 | 0.367802 | -0.633847 | 0.089529 |
| 7 | -0.203110 | 0.504909 | 0.016051 | 2.047894 | 3.947978 | -0.105809 | -0.891699 | -2.283474 | -0.394168 | 3.202955 | 0.720393 | 0.497918 | -0.659265 | 0.014837 |
| 8 | -0.186089 | 0.454411 | -0.238047 | 2.045116 | 3.660860 | -0.251945 | -0.904483 | -2.138275 | -1.225798 | 2.962659 | 0.756347 | 0.422399 | -0.794784 | -0.045474 |
| 9 | -0.292553 | 0.451394 | -0.383922 | 2.077637 | 14.496776 | -0.136514 | -1.136826 | -2.143701 | -1.429141 | 3.352161 | 0.555814 | 0.549724 | -0.665753 | -0.115536 |
And visualize them:
[21]:
sns.clustermap(mean_acts, xticklabels=mean_acts.columns, cmap='coolwarm', z_score=1)
plt.show()
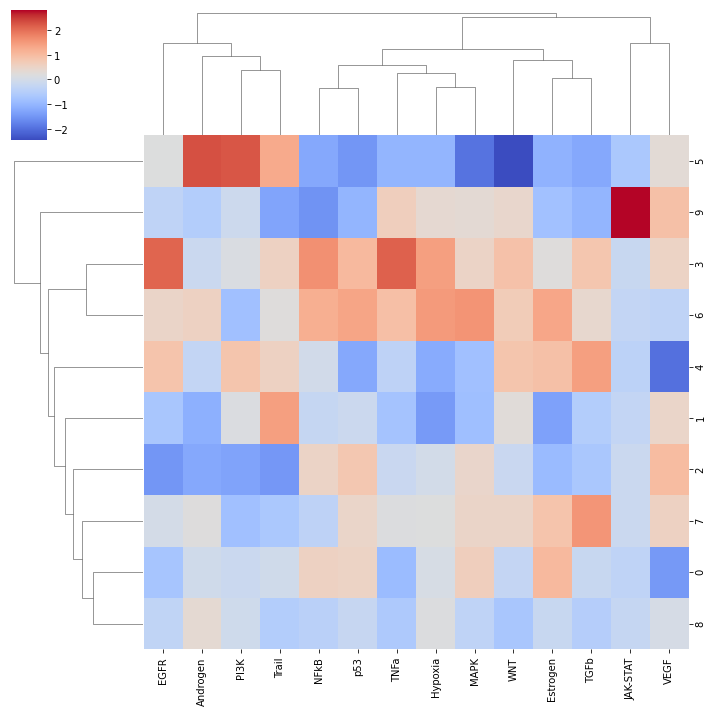
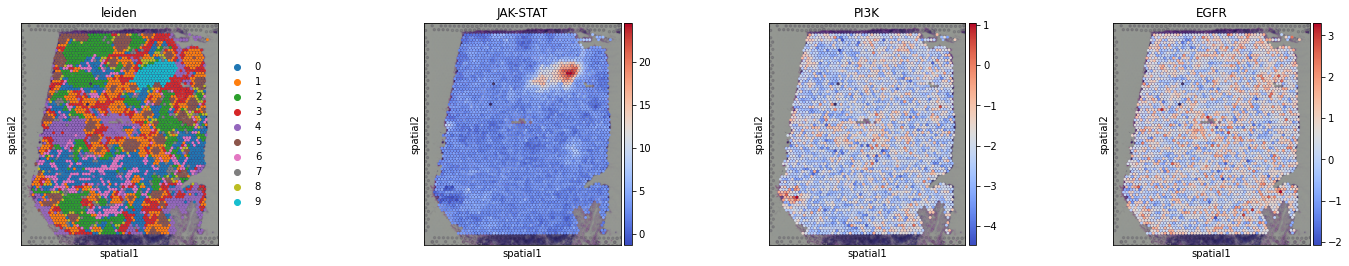
We can visualize again on the tissue some cluster-specific pathways:
[22]:
sc.pl.spatial(acts, color=['leiden', 'JAK-STAT', 'PI3K', 'EGFR'], cmap='coolwarm', size=1.5, wspace=0)
Functional enrichment of biological terms
Finally, we can also infer activities for general biological terms or processes.
MSigDB gene sets
The Molecular Signatures Database (MSigDB) is a resource containing a collection of gene sets annotated to different biological processes.
[23]:
msigdb = dc.get_resource('MSigDB')
msigdb
[23]:
| genesymbol | collection | geneset | |
|---|---|---|---|
| 0 | MSC | oncogenic_signatures | PKCA_DN.V1_DN |
| 1 | MSC | mirna_targets | MIR12123 |
| 2 | MSC | chemical_and_genetic_perturbations | NIKOLSKY_BREAST_CANCER_8Q12_Q22_AMPLICON |
| 3 | MSC | immunologic_signatures | GSE32986_UNSTIM_VS_GMCSF_AND_CURDLAN_LOWDOSE_S... |
| 4 | MSC | chemical_and_genetic_perturbations | BENPORATH_PRC2_TARGETS |
| ... | ... | ... | ... |
| 2407729 | OR2W5P | immunologic_signatures | GSE22601_DOUBLE_NEGATIVE_VS_CD8_SINGLE_POSITIV... |
| 2407730 | OR2W5P | immunologic_signatures | KANNAN_BLOOD_2012_2013_TIV_AGE_65PLS_REVACCINA... |
| 2407731 | OR52L2P | immunologic_signatures | GSE22342_CD11C_HIGH_VS_LOW_DECIDUAL_MACROPHAGE... |
| 2407732 | CSNK2A3 | immunologic_signatures | OCONNOR_PBMC_MENVEO_ACWYVAX_AGE_30_70YO_7DY_AF... |
| 2407733 | AQP12B | immunologic_signatures | MATSUMIYA_PBMC_MODIFIED_VACCINIA_ANKARA_VACCIN... |
2407734 rows × 3 columns
As an example, we will use the hallmark gene sets, but we could have used any other.
Note
To see what other collections are available in MSigDB, type: msigdb['collection'].unique().
We can filter by for hallmark:
[24]:
# Filter by hallmark
msigdb = msigdb[msigdb['collection']=='hallmark']
# Remove duplicated entries
msigdb = msigdb[~msigdb.duplicated(['geneset', 'genesymbol'])]
# Rename
msigdb.loc[:, 'geneset'] = [name.split('HALLMARK_')[1] for name in msigdb['geneset']]
msigdb
[24]:
| genesymbol | collection | geneset | |
|---|---|---|---|
| 11 | MSC | hallmark | TNFA_SIGNALING_VIA_NFKB |
| 149 | ICOSLG | hallmark | TNFA_SIGNALING_VIA_NFKB |
| 223 | ICOSLG | hallmark | INFLAMMATORY_RESPONSE |
| 270 | ICOSLG | hallmark | ALLOGRAFT_REJECTION |
| 398 | FOSL2 | hallmark | HYPOXIA |
| ... | ... | ... | ... |
| 878342 | FOXO1 | hallmark | PANCREAS_BETA_CELLS |
| 878418 | GCG | hallmark | PANCREAS_BETA_CELLS |
| 878512 | PDX1 | hallmark | PANCREAS_BETA_CELLS |
| 878605 | INS | hallmark | PANCREAS_BETA_CELLS |
| 878785 | SRP9 | hallmark | PANCREAS_BETA_CELLS |
7318 rows × 3 columns
Activity inference with Multivariate Linear Model
Like in the previous section, we will use mlm to infer activities:
[25]:
dc.run_mlm(mat=adata, net=msigdb, source='geneset', target='genesymbol', weight=None, verbose=True)
# Store them in a different key
adata.obsm['msigdb_mlm_estimate'] = adata.obsm['mlm_estimate'].copy()
adata.obsm['msigdb_mlm_pvals'] = adata.obsm['mlm_pvals'].copy()
Running mlm on mat with 4032 samples and 19813 targets for 50 sources.
100%|██████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████| 1/1 [00:04<00:00, 4.03s/it]
The obtained scores (t-values)(mlm_estimate) and p-values (mlm_pvals) are stored in the .obsm key:
[26]:
adata.obsm['msigdb_mlm_estimate'].iloc[:, 0:5]
[26]:
| ADIPOGENESIS | ALLOGRAFT_REJECTION | ANDROGEN_RESPONSE | ANGIOGENESIS | APICAL_JUNCTION | |
|---|---|---|---|---|---|
| AAACAAGTATCTCCCA-1 | 3.582305 | 11.334203 | 4.234747 | 0.137435 | 3.802077 |
| AAACAATCTACTAGCA-1 | 2.671094 | 12.194424 | 2.753398 | -0.224686 | 2.211782 |
| AAACACCAATAACTGC-1 | 3.254313 | 11.859533 | 4.187515 | -0.349350 | 3.922884 |
| AAACAGAGCGACTCCT-1 | 1.460420 | 11.618388 | 4.099222 | -0.313021 | 4.280679 |
| AAACAGCTTTCAGAAG-1 | 1.603691 | 12.120145 | 2.952333 | -0.271135 | 2.790807 |
| ... | ... | ... | ... | ... | ... |
| TTGTTTCACATCCAGG-1 | 2.417024 | 9.619124 | 4.501319 | 0.699741 | 3.429903 |
| TTGTTTCATTAGTCTA-1 | 3.775330 | 11.446894 | 4.151324 | 0.583313 | 5.161738 |
| TTGTTTCCATACAACT-1 | 2.932805 | 12.189088 | 3.406567 | -0.550669 | 4.736694 |
| TTGTTTGTATTACACG-1 | 1.151807 | 10.742589 | 2.760084 | -0.723917 | 3.925781 |
| TTGTTTGTGTAAATTC-1 | 1.686387 | 11.515235 | 3.075319 | -1.084208 | 3.733509 |
4032 rows × 5 columns
Visualization
Like in the previous sections, we will extract the activities from the adata object.
[27]:
acts = dc.get_acts(adata, obsm_key='msigdb_mlm_estimate')
acts
[27]:
AnnData object with n_obs × n_vars = 4032 × 50
obs: 'in_tissue', 'array_row', 'array_col', 'n_genes', 'n_genes_by_counts', 'total_counts', 'total_counts_mt', 'pct_counts_mt', 'leiden'
uns: 'spatial', 'log1p', 'hvg', 'pca', 'neighbors', 'leiden', 'leiden_colors'
obsm: 'spatial', 'X_pca', 'mlm_estimate', 'mlm_pvals', 'dorothea_mlm_estimate', 'dorothea_mlm_pvals', 'progeny_mlm_estimate', 'progeny_mlm_pvals', 'msigdb_mlm_estimate', 'msigdb_mlm_pvals'
Once extracted we can visualize them:
[28]:
sc.pl.spatial(acts, color='APICAL_JUNCTION', cmap='coolwarm', size=1.5)
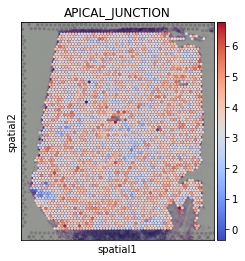
Exploration
Let us see the most variable terms:
[29]:
mean_acts = dc.summarize_acts(acts, groupby='leiden', min_std=0.35)
mean_acts
[29]:
| COAGULATION | DNA_REPAIR | E2F_TARGETS | EPITHELIAL_MESENCHYMAL_TRANSITION | G2M_CHECKPOINT | INTERFERON_ALPHA_RESPONSE | INTERFERON_GAMMA_RESPONSE | KRAS_SIGNALING_UP | MYC_TARGETS_V1 | OXIDATIVE_PHOSPHORYLATION | TNFA_SIGNALING_VIA_NFKB | UNFOLDED_PROTEIN_RESPONSE | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 0.952400 | 3.288563 | 1.090645 | 1.618884 | 2.080138 | 3.904741 | 5.837221 | 1.469192 | 18.919279 | 10.948023 | 3.336524 | 6.792207 |
| 1 | 0.507895 | 3.667026 | 1.263194 | 0.769735 | 2.176249 | 4.147588 | 5.594840 | 1.383072 | 19.796711 | 11.067318 | 2.634282 | 5.720784 |
| 2 | 0.713078 | 3.559376 | 0.962561 | 1.215796 | 1.934632 | 4.388570 | 5.846885 | 1.429250 | 19.889725 | 11.445617 | 3.115541 | 6.276733 |
| 3 | 1.784987 | 3.652838 | 0.858465 | 2.945634 | 1.832419 | 4.097078 | 6.311147 | 2.180722 | 20.106998 | 11.705830 | 4.497266 | 6.225229 |
| 4 | 1.605946 | 3.044741 | 0.842482 | 2.983389 | 1.584821 | 3.771497 | 5.217919 | 1.660632 | 18.754562 | 10.439170 | 3.145799 | 5.770047 |
| 5 | 0.669163 | 4.479780 | 5.285630 | 0.682815 | 4.448758 | 3.479510 | 4.705666 | 1.515177 | 21.911728 | 13.188506 | 1.886962 | 5.720539 |
| 6 | 1.797583 | 3.589422 | 0.699074 | 2.483427 | 1.931533 | 3.930642 | 6.288488 | 2.142133 | 19.284714 | 11.306396 | 4.472759 | 6.775112 |
| 7 | 1.766313 | 3.687238 | 0.970407 | 2.642560 | 2.070699 | 4.431295 | 6.062190 | 2.108777 | 19.949854 | 11.676679 | 3.498453 | 6.584166 |
| 8 | 0.750419 | 3.982705 | 2.575233 | 1.394051 | 2.954589 | 4.300572 | 5.852424 | 1.354083 | 21.482012 | 12.530923 | 3.024118 | 6.544428 |
| 9 | 0.693047 | 4.042583 | 2.055638 | 0.901221 | 2.784884 | 9.241279 | 9.130281 | 1.200511 | 21.893719 | 12.714638 | 2.457613 | 6.549897 |
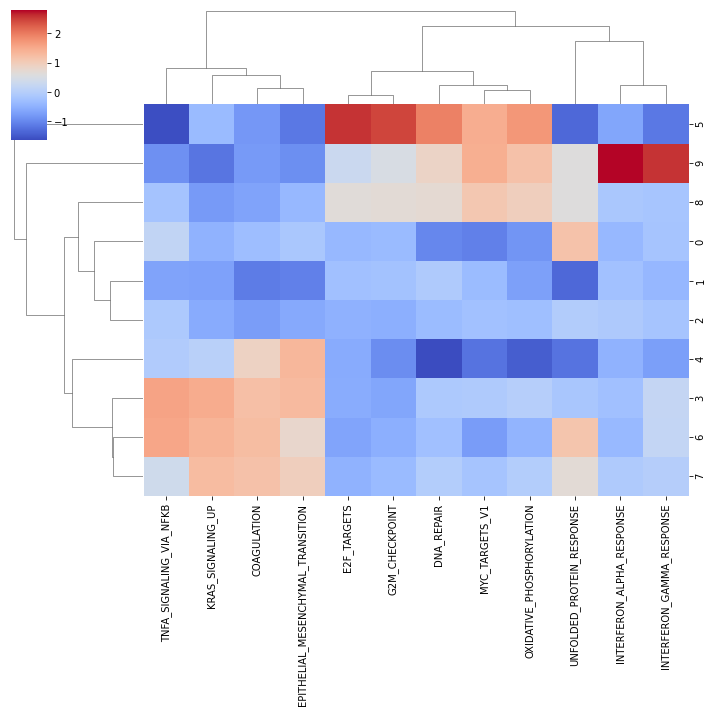
And visualize them:
[30]:
sns.clustermap(mean_acts, xticklabels=mean_acts.columns, cmap='coolwarm', z_score=1)
plt.show()
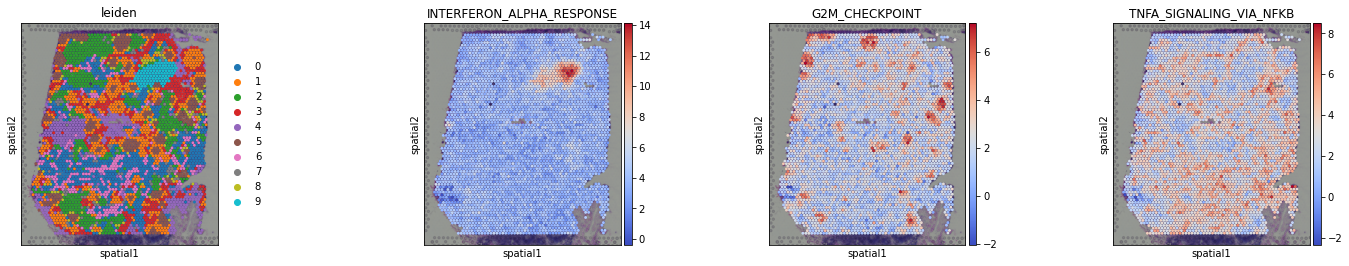
We can visualize again on the tissue some cluster-specific terms:
[31]:
sc.pl.spatial(acts, color=['leiden', 'INTERFERON_ALPHA_RESPONSE', 'G2M_CHECKPOINT', 'TNFA_SIGNALING_VIA_NFKB'], cmap='coolwarm', size=1.5, wspace=0)